MOC computation¶
If you would like to try to repeat examples from this notebook, you can download FESOM2 data and mesh. The data are quite heavy, about 15 Gb.
Link: https://swiftbrowser.dkrz.de/public/dkrz_c719fbc3-98ea-446c-8e01-356dac22ed90/PYFESOM2/
You have to download both archives (LCORE.tar and LCORE_MESH.tar) and extract them.
Alternative would be to use very light weight mesh that comes with pyfesom2 in the tests/data/pi-grid/ and example data on this mesh in tests/data/pi-results.
[13]:
import pyfesom2 as pf
import matplotlib.cm as cm
import matplotlib.pylab as plt
import numpy as np
[4]:
mesh = pf.load_mesh('/Users/koldunovn/PYHTON/DATA/LCORE_MESH/',
abg=[50, 15, -90])
/Users/koldunovn/PYHTON/DATA/LCORE_MESH/pickle_mesh_py3_fesom2
The usepickle == True)
The pickle file for FESOM2 exists.
The mesh will be loaded from /Users/koldunovn/PYHTON/DATA/LCORE_MESH/pickle_mesh_py3_fesom2
[6]:
data = pf.get_data('../../DATA/LCORE/', 'w', range(1950,1959), mesh, how="mean", compute=True )
Depth is None, 3d field will be returned
[28]:
data.shape
[28]:
(126858, 48)
Minimum nessesary input is mesh and one 3D field of w.
[34]:
lats, moc = pf.xmoc_data(mesh, data)
You get latitides and the MOC values (global by default). They can be used to plot the MOC using hofm_plot function:
[35]:
plt.figure(figsize=(10, 3))
pf.hofm_plot(mesh, moc, xvals=lats, maxdepth=7000, cmap=cm.seismic, levels = np.linspace(-30, 30, 51),
facecolor='gray')
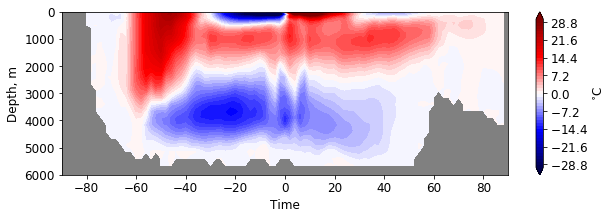
Increase the number of latitude bins:
[36]:
lats, moc = pf.xmoc_data(mesh, data, nlats=200)
[37]:
plt.figure(figsize=(10, 3))
pf.hofm_plot(mesh, moc, xvals=lats, maxdepth=7000, cmap=cm.seismic, levels = np.linspace(-30, 30, 51),
facecolor='gray')
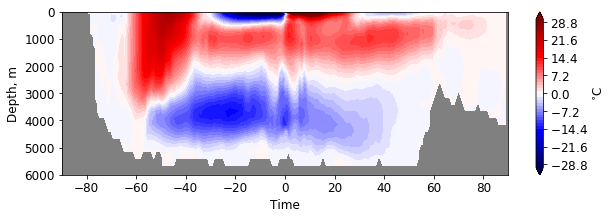
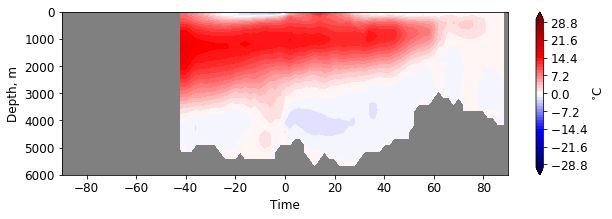
You can define the region for MOC computation.
[38]:
lats, moc = pf.xmoc_data(mesh, data, mask="Atlantic_MOC")
[39]:
plt.figure(figsize=(10, 3))
pf.hofm_plot(mesh, moc, xvals=lats, maxdepth=7000, cmap=cm.seismic, levels = np.linspace(-30, 30, 51),
facecolor='gray')
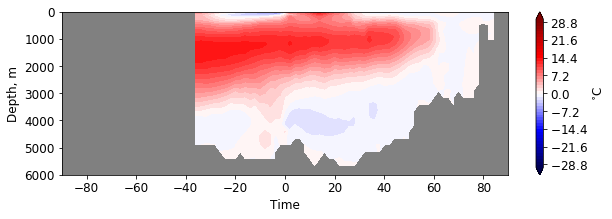
Regions are listed in documentation of pf.get_mask. Currently they are:
Ocean Basins:
"Atlantic_Basin"
"Pacific_Basin"
"Indian_Basin"
"Arctic_Basin"
"Southern_Ocean_Basin"
"Mediterranean_Basin"
"Global Ocean"
"Global Ocean 65N to 65S"
"Global Ocean 15S to 15N"
MOC Basins:
"Atlantic_MOC"
"IndoPacific_MOC"
"Pacific_MOC"
"Indian_MOC"
Nino Regions:
"Nino 3.4"
"Nino 3"
"Nino 4"
You can combine masks:
[49]:
mask1 = pf.get_mask(mesh, "Atlantic_Basin")
mask2 = pf.get_mask(mesh, "Arctic_Basin")
mask3 = mask1|mask2
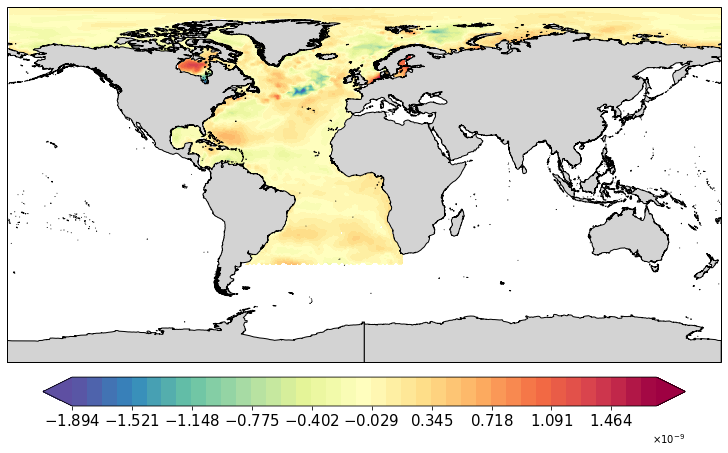
Check how the masked data looks like:
[50]:
pf.plot(mesh, data[:,0]*mask3)
[50]:
[<cartopy.mpl.geoaxes.GeoAxesSubplot at 0x11ed73ef0>]
You can then provide combined mask to xmoc_data:
[61]:
lats, moc = pf.xmoc_data(mesh, data, mask=mask3)
[62]:
plt.figure(figsize=(10, 3))
pf.hofm_plot(mesh, moc, xvals=lats, maxdepth=7000, cmap=cm.seismic, levels = np.linspace(-30, 30, 51),
facecolor='gray')
If you work with large meshes, it makes sence to compute some data beforehand and provide them to the function:
[63]:
meshdiag = pf.get_meshdiag(mesh)
el_area = meshdiag['elem_area'][:]
nlevels = meshdiag['nlevels'][:]-1
face_x, face_y = pf.compute_face_coords(mesh)
mask = pf.get_mask(mesh, 'Global Ocean')
[64]:
lats, moc = pf.xmoc_data(mesh, data, mask = mask,
el_area = el_area, nlevels=nlevels,
face_x=face_x, face_y=face_y)
[65]:
plt.figure(figsize=(10, 3))
pf.hofm_plot(mesh, moc, xvals=lats, maxdepth=7000, cmap=cm.seismic, levels = np.linspace(-30, 30, 51),
facecolor='gray')
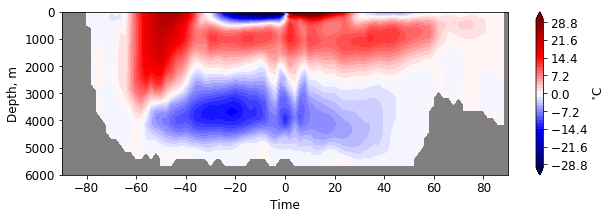
Compute AMOC at 26.5N¶
Open data as time series
[88]:
data = pf.get_data('../../DATA/LCORE/', 'w', range(1950,1959), mesh, how="ori", compute=False )
Depth is None, 3d field will be returned
[89]:
data
[89]:
- time: 9
- nod2: 126858
- nz: 48
- dask.array<chunksize=(1, 126858, 48), meta=np.ndarray>
Array Chunk Bytes 219.21 MB 24.36 MB Shape (9, 126858, 48) (1, 126858, 48) Count 27 Tasks 9 Chunks Type float32 numpy.ndarray - time(time)datetime64[ns]1950-12-31T23:15:00 ... 1958-12-31T23:15:00
- long_name :
- time
array(['1950-12-31T23:15:00.000000000', '1951-12-31T23:15:00.000000000', '1952-12-30T23:15:00.000000000', '1953-12-31T23:15:00.000000000', '1954-12-31T23:15:00.000000000', '1955-12-31T23:15:00.000000000', '1956-12-30T23:15:00.000000000', '1957-12-31T23:15:00.000000000', '1958-12-31T23:15:00.000000000'], dtype='datetime64[ns]')
- description :
- vertical velocity
- units :
- m/s
We have 9 time steps
[90]:
data.shape
[90]:
(9, 126858, 48)
Compute AMOC for each time step, increase the number of latitudes if you want to be preciselly at 26.5:
[109]:
moc = []
for i in range(data.shape[0]):
lats, moc_time = pf.xmoc_data(mesh, data[i,:,:], mask='Atlantic_MOC', nlats=361)
moc.append(moc_time)
Find which index corresponds to 26.5
[110]:
lats[233]
[110]:
26.5
Compute maximum at this point for each time step:
[113]:
amoc26 = []
for i in range(len(moc)):
amoc26.append(moc[i][233,:].max())
[114]:
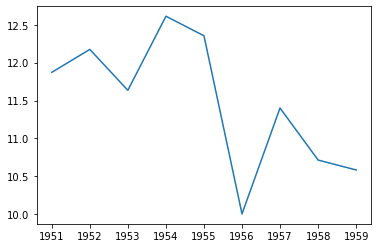
amoc26
[114]:
[11.871677395438851,
12.17447911245645,
11.634695012527837,
12.611884810155964,
12.354251356017224,
10.003526012070289,
11.40134476396998,
10.713783333512506,
10.583684577084185]
You can use time information from original dataset for plotting:
[116]:
plt.plot(data.time, amoc26)
[116]:
[<matplotlib.lines.Line2D at 0x11eb854a8>]
[ ]:
[ ]: